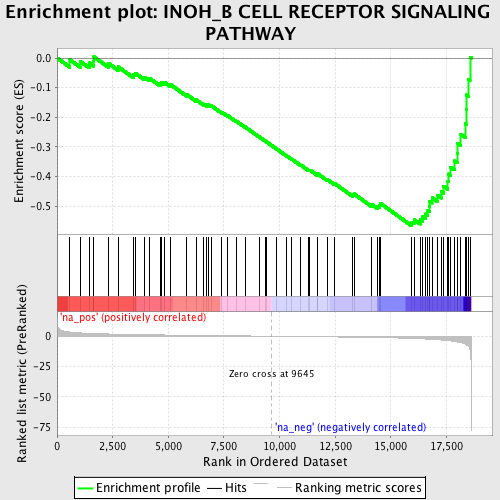
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | INOH_B CELL RECEPTOR SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.56724894 |
| Normalized Enrichment Score (NES) | -1.8677958 |
| Nominal p-value | 0.0014104373 |
| FDR q-value | 0.14576003 |
| FWER p-Value | 0.74 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | NFKBIE | 575 | 3.473 | -0.0059 | No | ||
| 2 | NFKBIB | 1055 | 2.697 | -0.0123 | No | ||
| 3 | PIK3R2 | 1444 | 2.324 | -0.0164 | No | ||
| 4 | GRB2 | 1642 | 2.197 | -0.0112 | No | ||
| 5 | CRK | 1653 | 2.190 | 0.0041 | No | ||
| 6 | ITPR3 | 2321 | 1.829 | -0.0187 | No | ||
| 7 | HRAS | 2740 | 1.631 | -0.0294 | No | ||
| 8 | SOS1 | 3421 | 1.389 | -0.0561 | No | ||
| 9 | GAB2 | 3505 | 1.363 | -0.0507 | No | ||
| 10 | IRS4 | 3939 | 1.231 | -0.0652 | No | ||
| 11 | PRKCD | 4150 | 1.173 | -0.0680 | No | ||
| 12 | MYD88 | 4630 | 1.054 | -0.0862 | No | ||
| 13 | FRS3 | 4679 | 1.042 | -0.0813 | No | ||
| 14 | SOS2 | 4831 | 1.008 | -0.0822 | No | ||
| 15 | DOK1 | 5087 | 0.949 | -0.0891 | No | ||
| 16 | PIK3R1 | 5830 | 0.769 | -0.1235 | No | ||
| 17 | SH2B3 | 6242 | 0.681 | -0.1408 | No | ||
| 18 | NFKB1 | 6591 | 0.610 | -0.1551 | No | ||
| 19 | GRB14 | 6729 | 0.586 | -0.1583 | No | ||
| 20 | SHC3 | 6799 | 0.569 | -0.1579 | No | ||
| 21 | FADD | 6921 | 0.544 | -0.1605 | No | ||
| 22 | PRKCA | 7407 | 0.444 | -0.1834 | No | ||
| 23 | MAP2K2 | 7680 | 0.391 | -0.1953 | No | ||
| 24 | KRAS | 8061 | 0.313 | -0.2135 | No | ||
| 25 | PRKCZ | 8446 | 0.236 | -0.2325 | No | ||
| 26 | BCAR1 | 9088 | 0.110 | -0.2663 | No | ||
| 27 | NRAS | 9380 | 0.052 | -0.2816 | No | ||
| 28 | MAP2K1 | 9431 | 0.042 | -0.2840 | No | ||
| 29 | PIK3CA | 9878 | -0.045 | -0.3077 | No | ||
| 30 | TRAF5 | 10299 | -0.129 | -0.3294 | No | ||
| 31 | CD79A | 10539 | -0.174 | -0.3410 | No | ||
| 32 | CD19 | 10950 | -0.264 | -0.3612 | No | ||
| 33 | CRADD | 11312 | -0.342 | -0.3782 | No | ||
| 34 | SRC | 11366 | -0.352 | -0.3785 | No | ||
| 35 | GRB7 | 11709 | -0.430 | -0.3939 | No | ||
| 36 | MAPK3 | 11722 | -0.435 | -0.3914 | No | ||
| 37 | PRKCE | 12144 | -0.521 | -0.4103 | No | ||
| 38 | BLNK | 12466 | -0.598 | -0.4233 | No | ||
| 39 | PRKCI | 13267 | -0.791 | -0.4607 | No | ||
| 40 | IRS1 | 13344 | -0.810 | -0.4590 | No | ||
| 41 | GRAP2 | 14115 | -1.018 | -0.4931 | No | ||
| 42 | GRB10 | 14406 | -1.105 | -0.5008 | No | ||
| 43 | ARAF | 14504 | -1.136 | -0.4978 | No | ||
| 44 | LAT | 14531 | -1.143 | -0.4910 | No | ||
| 45 | RAF1 | 15947 | -1.805 | -0.5542 | Yes | ||
| 46 | SYK | 16044 | -1.873 | -0.5459 | Yes | ||
| 47 | SHC1 | 16319 | -2.078 | -0.5457 | Yes | ||
| 48 | PRKCH | 16424 | -2.159 | -0.5357 | Yes | ||
| 49 | PIK3CD | 16562 | -2.267 | -0.5267 | Yes | ||
| 50 | TRAF6 | 16659 | -2.353 | -0.5149 | Yes | ||
| 51 | RELA | 16736 | -2.440 | -0.5014 | Yes | ||
| 52 | DOK2 | 16749 | -2.456 | -0.4843 | Yes | ||
| 53 | LCK | 16851 | -2.559 | -0.4713 | Yes | ||
| 54 | TRAF2 | 17084 | -2.837 | -0.4633 | Yes | ||
| 55 | ITPR1 | 17263 | -3.094 | -0.4506 | Yes | ||
| 56 | PRKCQ | 17373 | -3.270 | -0.4328 | Yes | ||
| 57 | NFKBIA | 17567 | -3.627 | -0.4171 | Yes | ||
| 58 | VAV1 | 17585 | -3.659 | -0.3916 | Yes | ||
| 59 | SH2D2A | 17679 | -3.861 | -0.3687 | Yes | ||
| 60 | FYN | 17854 | -4.233 | -0.3476 | Yes | ||
| 61 | LCP2 | 17979 | -4.636 | -0.3208 | Yes | ||
| 62 | CRKL | 17994 | -4.686 | -0.2877 | Yes | ||
| 63 | CBL | 18150 | -5.305 | -0.2578 | Yes | ||
| 64 | CD79B | 18344 | -6.466 | -0.2215 | Yes | ||
| 65 | TRAF3 | 18385 | -6.833 | -0.1744 | Yes | ||
| 66 | TRAF4 | 18399 | -6.988 | -0.1246 | Yes | ||
| 67 | TRAF1 | 18476 | -8.002 | -0.0710 | Yes | ||
| 68 | GAB1 | 18560 | -10.871 | 0.0030 | Yes |
